Restriction Enzyme (Restriction Endonuclease)

Restriction Enzyme (Restriction Endonuclease) Definition
- Restriction enzyme, also called restriction endonuclease, is a protein produced by bacteria that cleaves DNA at specific sites along the molecule.
- Restriction endonucleases cut the DNA double helix in very precise ways. It cleaves DNA into fragments at or near specific recognition sites within the molecule known as restriction sites.
- They have the capacity to recognize specific base sequences on DNA and then to cut each strand at a given place. Hence, they are also called as ‘molecular scissors’.
Source of Restriction Enzymes
- The natural source of restriction endonucleases are bacterial cells.
- These enzymes are called restriction enzymes because they restrict infection of bacteria by certain viruses (i.e., bacteriophages), by degrading the viral DNA without affecting the bacterial DNA. Thus, their function in the bacterial cell is to destroy foreign DNA that might enter the cell.
- The restriction enzyme recognizes the foreign DNA and cuts it at several sites along the molecule.
- Each bacterium has its own unique restriction enzymes and each enzyme recognizes only one type of sequence.
Recognition Sites
- The DNA sequences recognized by restriction enzymes are called palindromes. Palindromes are the base sequences that read the same on the two strands but in opposite directions.
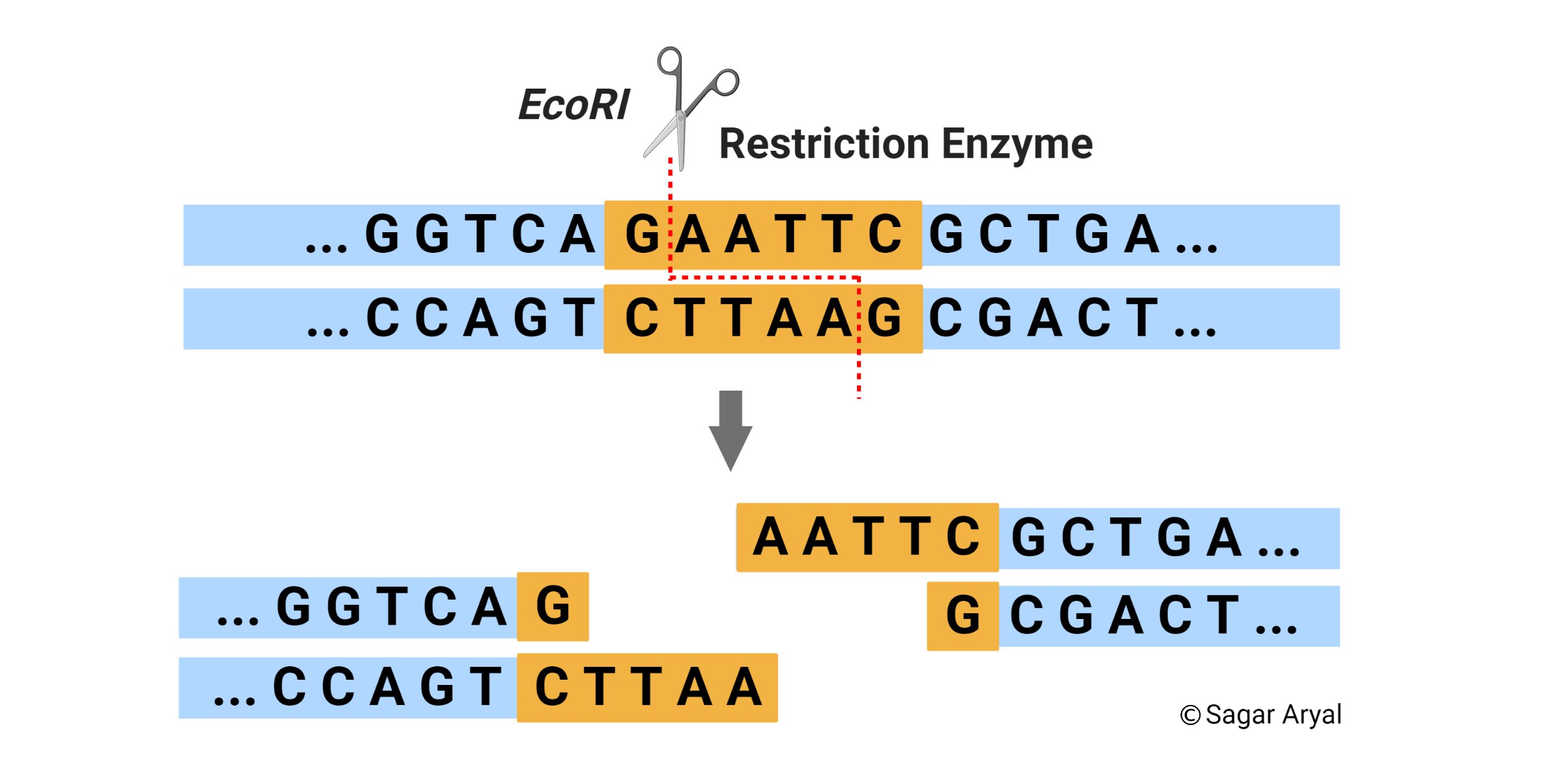
For example, if the sequence on one strand is GAATTC read in 5’→3′ direction, the sequence on the opposite strand is CTTAAG read in the 3’→5′ direction, but when both strands are read in the 5’→ 3′ direction the sequence is the same. The palindrome appears accordingly —
In addition, there is a point of symmetry within the palindrome. In the example, this point is in the center between the AT/AT.
- The value of restriction enzymes is that they make cuts in the DNA molecule around this point of symmetry.
- Some enzymes cut straight across the molecule at the symmetrical axis producing blunt ends.
- Of more value, however, are the restriction enzymes that cut between the same two bases away from the point of symmetry on two strands, thus, producing a staggering break.
Mechanism of Cleavage of Restriction Enzymes
When a restriction endonuclease recognizes a particular sequence, it snips through the DNA molecule by catalyzing the hydrolysis (splitting of a chemical bond by addition of a water molecule) of the bond between adjacent nucleotides. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix.
Types of Restriction Enzymes
Traditionally, four types of restriction enzymes are recognized, designated I, II, III, and IV, which differ primarily in structure, cleavage site, specificity, and cofactors.
- Type I enzymes cleave at sites remote from a recognition site; require both ATP and S-adenosyl-L-methionine to function; multifunctional protein with both restriction and methylase activities.
- Type II enzymes cleave within or at short specific distances from a recognition site; most require magnesium; single function (restriction) enzymes independent of methylase.
- Type III enzymes cleave at sites a short distance from a recognition site; require ATP (but do not hydrolyze it); S-adenosyl-L-methionine stimulates the reaction but is not required; it exists as part of a complex with a modification methylase.
- Type IV enzymes target modified DNA, e.g. methylated, hydroxymethylated and glucosyl-hydroxymethylated DNA.
Nomenclature of Restriction Enzymes
Since their discovery in the 1970s, many restriction enzymes have been identified while Type II restriction enzymes have been characterized.
Each enzyme is named after the bacterium from which it was isolated, using a naming system based on bacterial genus, species and strain. For example, the name of the EcoRI restriction enzyme was derived as:
E – Escherichia: Genus
co- coli: specific species
R- RY13: strain
I- First identified: order of identification in the bacterium
Applications of Restriction Enzymes
- Restriction enzymes can be isolated from bacterial cells and used in the laboratory to manipulate fragments of DNA, such as those that contain genes; for this reason, they are indispensable tools of recombinant DNA technology (genetic engineering).
- The most useful aspect of restriction enzymes is that each enzyme recognizes the same unique base sequence regardless of the source of the DNA. It means that these enzymes establish fixed landmarks along an otherwise very regular DNA molecule. This allows dividing a long DNA molecule into fragments that can be separated from each other by size with the technique of gel electrophoresis.
- Each fragment, thus generated, are also available for further analysis, including the sequencing.
- One value of cutting DNA molecule up into discrete fragments is being able to locate a particular gene on the fragment where it resides which is done by the general technique of Southern blotting.
Some examples of Restriction Enzymes
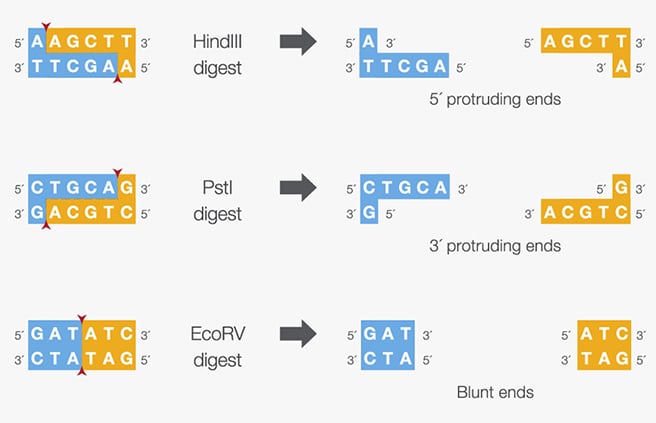
Figure: Sticky or protruding ends (5′ or 3′) or blunt ends produced by specific restriction enzymes.
Source: Thermo Fisher Scientific
- One of the most popular restriction enzymes is called EcoRI from E. coli (bacterium).
- Hundreds of other restriction enzymes with different sequence specificities have been isolated from several bacteria and are commercially available.
| Enzyme | Obtained from | Recognition Sequence |
| EcoRI | Escherichia coli | 5’GAATTC 3’CTTAAG |
| EcoRII | Escherichia coli | 5’CCWGG 3’GGWCC |
| BamHI | Bacillus amyloliquefaciens | 5’GGATCC 3’CCTAGG |
| HindIII | Haemophilus influenzae | 5’AAGCTT 3’TTCGAA |
References
- Verma, P. S., & Agrawal, V. K. (2006). Cell Biology, Genetics, Molecular Biology, Evolution & Ecology (1 ed.). S .Chand and company Ltd.
- Klug, W. S., & Cummings, M. R. (2003). Concepts of genetics. Upper Saddle River, N.J: Prentice Hall.
- https://www.britannica.com/science/restriction-enzyme
- https://international.neb.com/products/restriction-endonucleases/restriction-endonucleases
- https://en.wikipedia.org/wiki/Restriction_enzyme
- http://www.bio.miami.edu/dana/dox/restrictionenzymes.html
Restriction Enzyme (Restriction Endonuclease)
About Author
Sagar Aryal is a microbiologist and a scientific blogger. He is doing his Ph.D. at the Central Department of Microbiology, Tribhuvan University, Kathmandu, Nepal. He was awarded the DAAD Research Grant to conduct part of his Ph.D. research work for two years (2019-2021) at Helmholtz-Institute for Pharmaceutical Research Saarland (HIPS), Saarbrucken, Germany. Sagar is interested in research on actinobacteria, myxobacteria, and natural products. He is the Research Head of the Department of Natural Products, Kathmandu Research Institute for Biological Sciences (KRIBS), Lalitpur, Nepal. Sagar has more than ten years of experience in blogging, content writing, and SEO. Sagar was awarded the SfAM Communications Award 2015: Professional Communicator Category from the Society for Applied Microbiology (Now: Applied Microbiology International), Cambridge, United Kingdom (UK). Sagar is also the ASM Young Ambassador to Nepal for the American Society for Microbiology since 2023 onwards.
4 thoughts on “Restriction Enzyme (Restriction Endonuclease)”
Djamel Achira
Very interesting lecture. Reply
Sheik subhani
Very impressive matter on restriction enzymes need some deep more key notes like sequence of methylation modification system specific in type III restriction endonucleases
Develop exonucleases functions too
Thankq
subhanisheik MSc Microbiology Andhra university Visakhapatnam Reply
Sheryar Khan
excellent lecture ,much information about restriction enzyme Reply
Habibullah
Good lecture regarding the restrication enzyme I have never seen such a comprhensive information Reply
Leave a Comment Cancel reply
Topics / Categories

- Agricultural Microbiology (13)
- Bacteriology (124)
- Basic Microbiology (61)
- Biochemical Test (114)
- Biochemistry (165)
- Bioinformatics (23)
- Biology (209)
- Biotechnology (31)
- Cell Biology (107)
- Culture Media (67)
- Difference Between (89)
- Diseases (32)
- Environmental Microbiology (7)
- Epidemiology (23)
- Food Microbiology (51)
- Genetics (79)
- Human Anatomy and Physiology (75)
- Immunology (115)
- Instrumentation (64)
- Microscopy (27)
- Molecular Biology (78)
- Mycology (33)
- Parasitology (26)
- Pharmacology (14)
- Phycology (2)
- Protocols (9)
- Research Methodology (20)
- Staining (29)
- Syllabus (18)
- Virology (50)
- Mycorrhiza: Definition, Types and Significances
- Marine Ecosystem and its Abiotic and Biotic Components
- Homeostasis: Definition, Types, Examples, Applications
- Neutrophils: Definition, Structure, Count, Range, Functions
- Whole Transcriptome Sequencing (WTS) / Total RNA-Seq





